Bell, CG;
Teschendorff, AE;
Rakyan, VK;
Maxwell, AP;
Beck, S;
Savage, DA;
(2010)
Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus.
BMC Medical Genomics
, 3
, Article 33. 10.1186/1755-8794-3-33.

Preview |
PDF
1755-8794-3-33.pdf Download (507kB) |
![[thumbnail of Additional file 1: Supplementary Figure S1. Distribution of CpG sites on Infinium platform. (Top, Left) Chromosome Distribution of CpG sites. (Top, Right) Distance to Transcription Start Site of CpG Locus. (Bottom, Left) Interrogated CpG Islands: near ge]](https://discovery.ucl.ac.uk/129867/2.hassmallThumbnailVersion/1755-8794-3-33-s1.png) 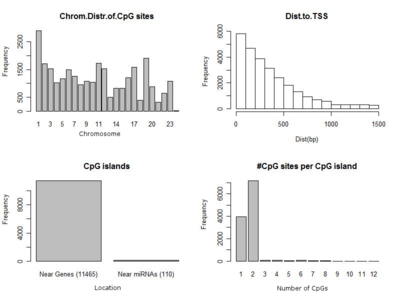 Preview |
Other (Additional file 1: Supplementary Figure S1. Distribution of CpG sites on Infinium platform. (Top, Left) Chromosome Distribution of CpG sites. (Top, Right) Distance to Transcription Start Site of CpG Locus. (Bottom, Left) Interrogated CpG Islands: near ge)
1755-8794-3-33-s1.png Download (133kB) |
|
Excel Spreadsheet (Additional file 2: Supplementary Table S1. List of CpGs with q value < 0.15. Column headings are IlmnID = Illumina CpG ID, Gene_ID = Entrez Gene ID, Symbol = Gene Symbol, coef = coefficient, exp(coef) = exponential of coefficient, se(coef) = standard err)
1755-8794-3-33-s2.xls Download (108kB) |
Abstract
Background: Diabetic nephropathy is a serious complication of diabetes mellitus and is associated with considerable morbidity and high mortality. There is increasing evidence to suggest that dysregulation of the epigenome is involved in diabetic nephropathy. We assessed whether epigenetic modification of DNA methylation is associated with diabetic nephropathy in a case-control study of 192 Irish patients with type 1 diabetes mellitus (T1D). Cases had T1D and nephropathy whereas controls had T1D but no evidence of renal disease.Methods: We performed DNA methylation profiling in bisulphite converted DNA from cases and controls using the recently developed Illumina Infinium (R) HumanMethylation27 BeadChip, that enables the direct investigation of 27,578 individual cytosines at CpG loci throughout the genome, which are focused on the promoter regions of 14,495 genes.Results: Singular Value Decomposition (SVD) analysis indicated that significant components of DNA methylation variation correlated with patient age, time to onset of diabetic nephropathy, and sex. Adjusting for confounding factors using multivariate Cox-regression analyses, and with a false discovery rate (FDR) of 0.05, we observed 19 CpG sites that demonstrated correlations with time to development of diabetic nephropathy. Of note, this included one CpG site located 18 bp upstream of the transcription start site of UNC13B, a gene in which the first intronic SNP rs13293564 has recently been reported to be associated with diabetic nephropathy.Conclusion: This high throughput platform was able to successfully interrogate the methylation state of individual cytosines and identified 19 prospective CpG sites associated with risk of diabetic nephropathy. These differences in DNA methylation are worthy of further follow-up in replication studies using larger cohorts of diabetic patients with and without nephropathy.
| Type: | Article |
|---|---|
| Title: | Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus |
| Open access status: | An open access version is available from UCL Discovery |
| DOI: | 10.1186/1755-8794-3-33 |
| Publisher version: | http://dx.doi.org/10.1186/1755-8794-3-33 |
| Language: | English |
| Additional information: | © 2010 Bell et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
| Keywords: | Susceptibility genes, Kidney-disease, Expression, Methylome, Risk, Hyperglycemia, Association, Epigenetics, Variants, Pathway |
| UCL classification: | UCL UCL > Provost and Vice Provost Offices > School of Life and Medical Sciences UCL > Provost and Vice Provost Offices > School of Life and Medical Sciences > Faculty of Medical Sciences UCL > Provost and Vice Provost Offices > School of Life and Medical Sciences > Faculty of Medical Sciences > Cancer Institute UCL > Provost and Vice Provost Offices > School of Life and Medical Sciences > Faculty of Medical Sciences > Cancer Institute > Research Department of Cancer Bio |
| URI: | https://discovery.ucl.ac.uk/id/eprint/129867 |
Archive Staff Only
 |
View Item |


