Ellingford, JM;
Campbell, C;
Barton, S;
Bhaskar, S;
Gupta, S;
Taylor, RL;
Sergouniotis, PI;
... Black, GCM; + view all
(2017)
Validation of copy number variation analysis for next-generation sequencing diagnostics.
European Journal of Human Genetics
, 25
(6)
pp. 719-724.
10.1038/ejhg.2017.42.

Preview |
Text
Michaelides_Ellingford-et-al-revised-2.pdf - Accepted Version Download (776kB) | Preview |
Preview |
Text (Supplementary Table)
Michaelides_Table 1 - Key factor analysis - REVISED.pdf - Accepted Version Download (357kB) | Preview |
![[thumbnail of Figure - 1]](https://discovery.ucl.ac.uk/1550022/12.hassmallThumbnailVersion/Michaelides_Figure-1-study-design-GREY.jpg)  Preview |
Image (Figure - 1)
Michaelides_Figure-1-study-design-GREY.jpg - Accepted Version Download (207kB) | Preview |
![[thumbnail of Figure - 2]](https://discovery.ucl.ac.uk/1550022/17.hassmallThumbnailVersion/Michaelides_Figure-2-FBN1-GREY.jpg)  Preview |
Image (Figure - 2)
Michaelides_Figure-2-FBN1-GREY.jpg - Accepted Version Download (122kB) | Preview |
![[thumbnail of Figure - 3]](https://discovery.ucl.ac.uk/1550022/22.hassmallThumbnailVersion/Michaelides_Figure-3-dx-and-simulations-GREY.jpg) 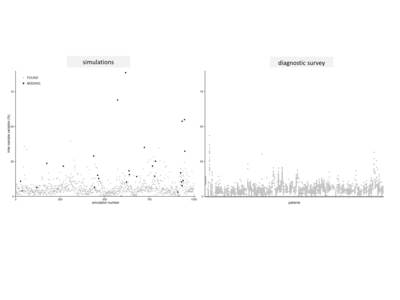 Preview |
Image (Figure - 3)
Michaelides_Figure-3-dx-and-simulations-GREY.jpg - Accepted Version Download (167kB) | Preview |
Abstract
Although a common cause of disease, copy number variants (CNVs) have not routinely been identified from next-generation sequencing (NGS) data in a clinical context. This study aimed to examine the sensitivity and specificity of a widely used software package, ExomeDepth, to identify CNVs from targeted NGS data sets. We benchmarked the accuracy of CNV detection using ExomeDepth v1.1.6 applied to targeted NGS data sets by comparison to CNV events detected through whole-genome sequencing for 25 individuals and determined the sensitivity and specificity of ExomeDepth applied to these targeted NGS data sets to be 100% and 99.8%, respectively. To define quality assurance metrics for CNV surveillance through ExomeDepth, we undertook simulation of single-exon (n=1000) and multiple-exon heterozygous deletion events (n=1749), determining a sensitivity of 97% (n=2749). We identified that the extent of sequencing coverage, the inter- and intra-sample variability in the depth of sequencing coverage and the composition of analysis regions are all important determinants of successful CNV surveillance through ExomeDepth. We then applied these quality assurance metrics during CNV surveillance for 140 individuals across 12 distinct clinical areas, encompassing over 500 potential rare disease diagnoses. All 140 individuals lacked molecular diagnoses after routine clinical NGS testing, and by application of ExomeDepth, we identified 17 CNVs contributing to the cause of a Mendelian disorder. Our findings support the integration of CNV detection using ExomeDepth v1.1.6 with routine targeted NGS diagnostic services for Mendelian disorders. Implementation of this strategy increases diagnostic yields and enhances clinical care.
| Type: | Article |
|---|---|
| Title: | Validation of copy number variation analysis for next-generation sequencing diagnostics |
| Open access status: | An open access version is available from UCL Discovery |
| DOI: | 10.1038/ejhg.2017.42 |
| Publisher version: | http://doi.org/10.1038/ejhg.2017.42 |
| Language: | English |
| Additional information: | This version is the author accepted manuscript. For information on re-use, please refer to the publisher’s terms and conditions. |
| UCL classification: | UCL UCL > Provost and Vice Provost Offices > School of Life and Medical Sciences UCL > Provost and Vice Provost Offices > School of Life and Medical Sciences > Faculty of Brain Sciences UCL > Provost and Vice Provost Offices > School of Life and Medical Sciences > Faculty of Brain Sciences > Institute of Ophthalmology |
| URI: | https://discovery.ucl.ac.uk/id/eprint/1550022 |
Archive Staff Only
 |
View Item |


